
Jared K. Slone
Houston-based scientist specializing in structural bioinformatics. My research interests center around using mathematical modeling and machine learning to solve difficult problems in biology.
Current Affiliations




Houston-based scientist specializing in structural bioinformatics. My research interests center around using mathematical modeling and machine learning to solve difficult problems in biology.
Current Affiliations



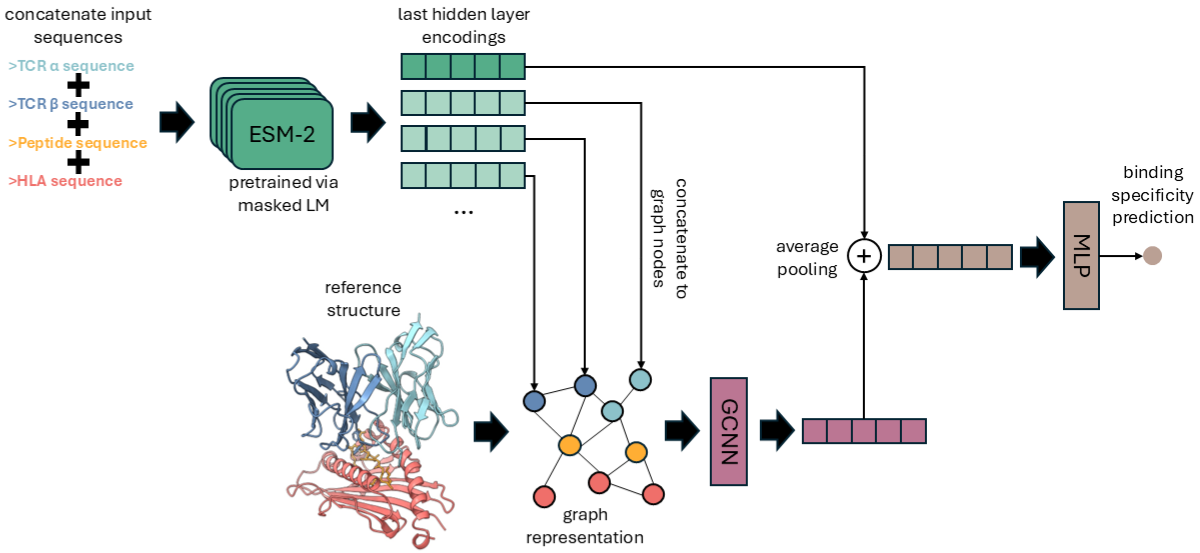
Strong binding between T cell receptors (TCRs) and peptide–HLA (pHLA) complexes plays a key role in the adaptive immune response. Accurately predicting which TCRs will bind to which pHLAs is central to advancing personalized immunotherapies such as TCR-T. Most existing ML approaches to TCR–pHLA binding prediction rely only on amino acid sequences, limiting their ability to capture the full complexity of binding interactions. Recent advances in protein structure modeling provide a unique opportunity to integrate 3D structural information into ML pipelines. We developed STAG-LLM, a multimodal model that combines protein language models with geometric deep learning to leverage both sequence data and computationally generated 3D structures of TCR–pHLA complexes in making predictions. Our results show that STAG-LLM outperforms existing methods, even when trained on datasets three times smaller. To further validate our model, we conduct in vitro alanine scanning experiments for four peptides and demonstrate a correlation with the attention weights learned by our model and in vitro results. Our findings demonstrate that structure-based approaches can meaningfully improve prediction of TCR–pHLA specificity.
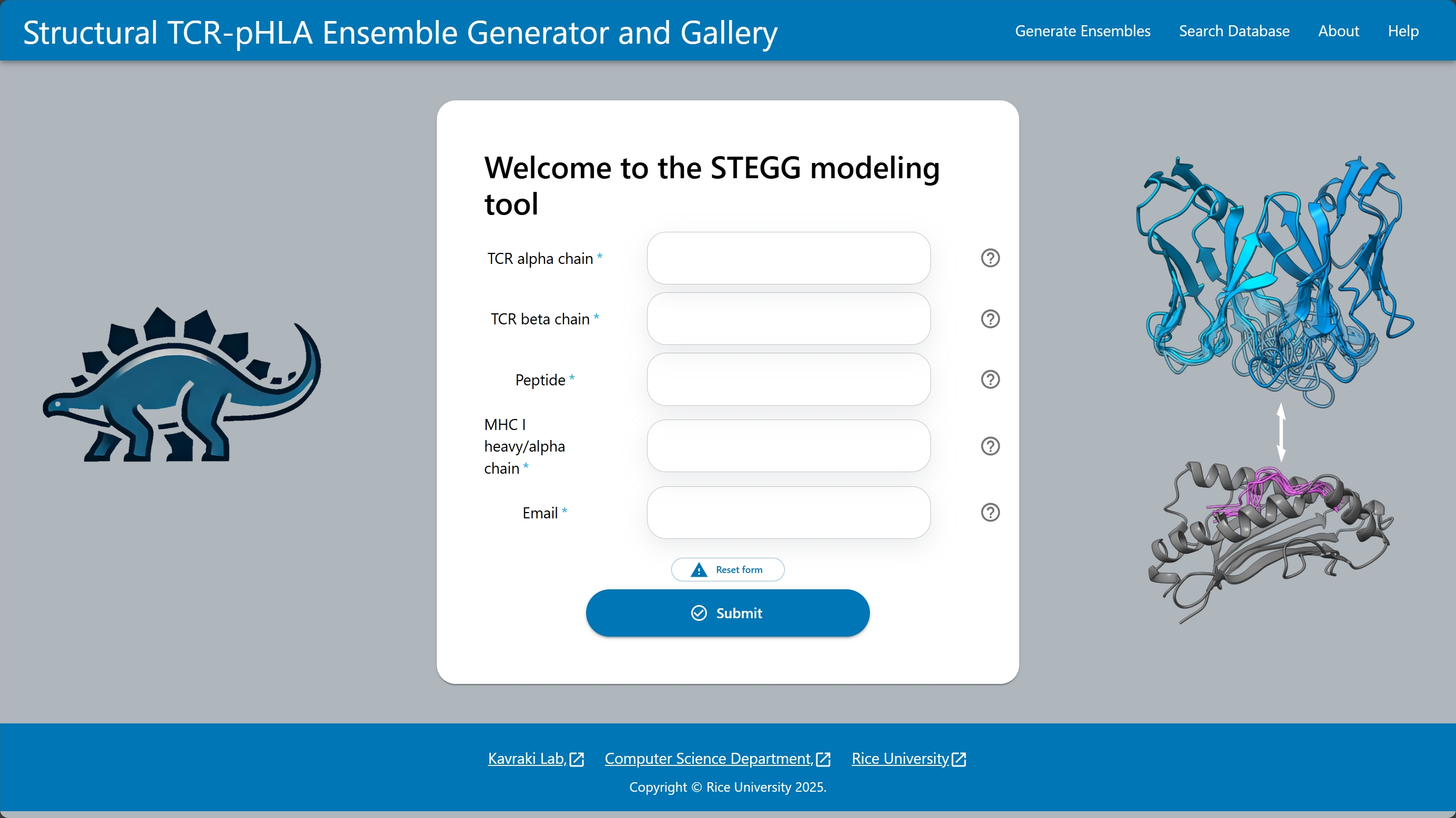
Understanding how T cell receptors (TCRs) recognize peptides presented by class-I major histocompatibility complexes (pMHCs) is a central challenge in immunology, with implications for infectious disease, autoimmunity, and personalized cancer immunotherapies. While recent 3D prediction advances have improved modeling, current methods fail to capture the flexible nature of TCR-pMHC interactions. Due to flexibility in TCR CDR loops and peptides, these dynamic interactions adopt a range of binding modes not represented by single static structures. We introduce STEGG: Structural TCR-pMHC Ensemble Generator and Gallery, a computational framework for generating structural ensembles from sequences. STEGG generates distinct low-energy 3D conformations, capturing the flexibility of TCRs, pMHCs, and their interactions. STEGG utilizes a domain-specific sampling algorithm for efficient modeling. We show that STEGG recovers conformations with low RMSD to experimentally solved structures and finds diverse, biologically relevant binding modes. We provide a publicly available database of structural ensembles. By going beyond static generation, STEGG presents a new way for studying TCR-pMHC interactions.
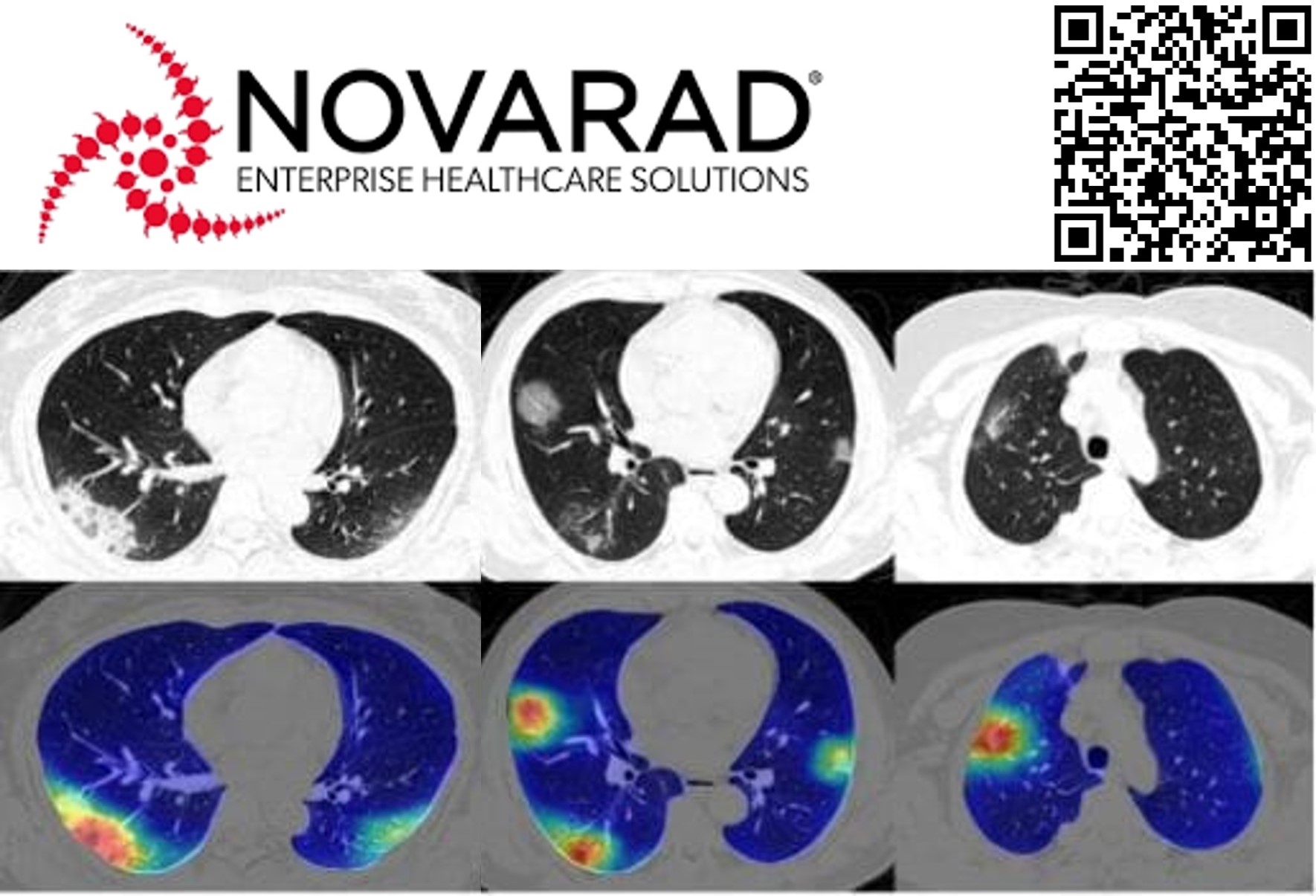
To address the high false-negative rates of PCR testing and the interpretation bottlenecks of manual radiology, we developed a computer vision solution for detecting COVID-19 in chest CT scans. Utilizing transfer learning, we fine-tuned an adapted ResNet-50 architecture on a dataset of 110 covid-positive CT scans and hundreds of pre-covid scans, including scans exhibiting other lung pathologies like COPD. The resulting model demonstrated robust performance across diverse clinical settings, achieving a sensitivity of 0.99 and a specificity of 0.92 on an external testing dataset of 438 scans. The system was deployed via a secure cloud infrastructure to ensure rapid, scalable access for providers. This project achieved widespread clinical adoption, being utilized in over 15 states and several countries to triage patients. By automating detection and quantifying lung involvement, this tool successfully augmented radiologist capabilities, offering a high-accuracy diagnostic aid during the global pandemic.
Jared K. Slone, Alexander W. Bock, Maurício Menegatti Rigo, Chengxuan Zou, Alexander Reuben, Lydia E. Kavraki.
Preprint / SSRN.
Jared K. Slone, Minying Zhang, Peixin Jiang, Amanda Montoya, Emily Bontekoe, Barbara Nassif Rausseo, Alexandre Reuben, Lydia E. Kavraki.
Computational and Structural Biotechnology Journal, 27, 3885-3896.
Jared K. Slone, Anja Conev, Maurício Menegatti Rigo, Alexandre Reuben, Lydia E. Kavraki.
IEEE Transactions on Computational Biology and Bioinformatics.
Amanda S Montoya, Peixin Jiang, Yulia Shulga, Sarah Forward, Emane Rose Assita, Emily G Bontekoe, Minying Zhang, Jared K. Slone, Ludovica La Posta, Eleonora Dondossola, Lydia Kavraki, Jianjun J Zhang, Trevor Brown, John V Heymach, Sheldon Kwok, Alexandre Reuben
Journal for ImmunoTherapy of Cancer, 13 (Suppl 2).
Amanda S Montoya, Meredith Frank, Peixin Jiang, Minying Zhang, Hui Nie, Emily Bontekoe, Jared K. Slone, Ludovica La Posta, Kavya L Singampalli, Desh Deepak Dixit, Weihong Xiao, Tina Cascone, Jianjun Zhang, Eleonora Dondossola, Lydia Kavraki, Maura Gillison, Pamela L Wenzel, Peter B Lillehoj, John V Heymach, Alexandre Reuben
Journal for ImmunoTherapy of Cancer, 12 (Suppl 2), 216.
Emily Bontekoe, Minying Zhang, Peixin Jiang, Amanda Montoya, Jared K. Slone, Ludovica La Posta, Jianjun Zhang, Cassian Yee, Eleonora Dondossola, Lydia Kavraki, Gregory Lizee, John V Heymach, Alexandre Reuben
Journal for ImmunoTherapy of Cancer, 12 (Suppl 2), 358.
Sarah Hall-Swan, Jared Slone, Maurício Menegatti Rigo, Dinler A. Antunes, Gregory Lizée, Lydia E. Kavraki.
Frontiers in Immunology, 14, 1108303.
Mark Hughes, Amy Eubanks, Jared Slone.
2021 Fall Western Sectional Meeting (American Mathematical Society).
Mark Hughes, Amy Eubanks, Jared Slone.
2021 Fall Central Sectional Meeting (American Mathematical Society).
Margaret C Lund, Marylesa M Howard, Chelsey E Noorda, Jared K. Slone.
Technical Report, Nevada National Security Site (NNSS), North Las Vegas, NV.


Biomedical Informatics and Data Science
Graduate Research Seminar, Rice University
Brigham Young University
Brigham Young University
Roy High School